|
|
|
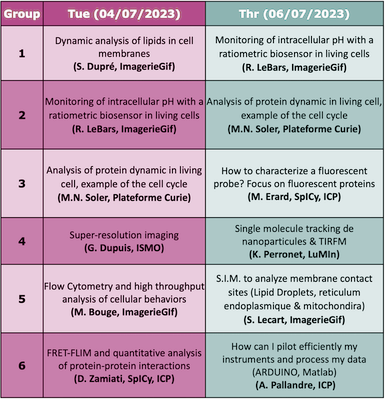
Hands-onRotation of the Hands-on:
Brief description
Marie Erard (ICP) Characterization of spectroscopic properties of a probe
A few key words on methods :
Location (adress and meeting point): Institute of Physical Chemistry, building 350 on the Orsay campus
Dounia Zamiati (ICP) Detection of FRET by FLIM to probe protein interactions at membrane contact sitesForster resonance energy transfer (FRET) between two fluorescent proteins (a donor and an acceptor) fused to proteins of interest is an efficient approach to monitor their interaction. We assess the FRET efficiency with the direct measurement of donor fluorescence lifetime (FLIM). This hands-on will present how to design such a FRET-FLIM experiment to monitor membrane contact sites. We will discuss the choice of FPs, the required controls and the data processing. A few key words on methods :
Location (adress and meeting point): Institute of Physical Chemistry, building 350 on the Orsay campus Karen Perronet (LuMIn) 2D single-particle tracking using Total Internal Reflection Fluorescence MicroscopyThe goals of this labwork are :
A few key words on methods :
Location (adress and meeting point): ENS Paris-Scalay 4 avenue des Sciences 91190 Gif sur Yvette – wait in the main lobby.
Antoine Pallandre Microfluidic and optical detectionThis practical training will allow you to start learning about microfluidic. Interfacing with Maltlab software to perform synchronization of optics, fluidic and electrical instruments will be explained in details. We will also build a microfluidic chip from a 3D printer before using it for microscopic observations. The use of different cameras (CMOS, CDD and modified webcam) will help you to better determine your need for your future microfluidic experiments. A few key words on methods : coding, microscopy, 3D printing, fluorescence, microfluidic and data acquisition
Location (adress and meeting point): Institute of Physical Chemistry, building 350 on the Orsay campus
Marie-Noëlle Soler (Institut Curie Centre de Recherche) Proteins recruitment after DNA damages on living cells, imaging and analysis
After DNA damages with 2 different lasers wavelengths, an acquisition timelapse allow to measure the recruitment of PARP2 protein in the damaged region. A few key words on methods :
Location (adress and meeting point): Institut Curie, Centre de Recherche sur le Centre Universitaire, building 110, Rue Henri Becquerel, 91401 Orsay Sandrine Lecart (I2BC) Investigation of endolplasmic reticulum – mitochondria contact sites using structured illumination microscopy (S.I.M.)Mitochondria–endoplasmic reticulum contact sites are unique environments within cells where important processes occurs (calcium homeostasis / lipid metabolism / autophagosome formation …). It is essential to understand its structure and identify the proteins recruited at those specific locations. To investigate this question a high level of resolution is required. In this workshop we will use a super resolution light microscopy technic (SIM) to image the ER-Mitochondria contact sites. A few key words on methods :
Location (adress and meeting point): 1 Avenue de la terrasse, building 21 (ground floor of the Hall), 91198 Gif-sur-Yvette cedex
Monitoring of intracellular pH with a ratiometric biosensor in living cellsThe legume-rhizobium symbiosis is characterized by the formation of symbiotic root nodules, in which the bacteria differentiate into nitrogen-fixing bacteroids. In order to identify novel features of terminal bacteroid differentiation, we tested on free-living bacteria and bacteroids undergoing TBD (Sinorhizobium meliloti in Medicago nodules) a novel genetically encoded ratiometric fluorescent biosensor (pHP, derived from GFP) to estimate intracellular pH. A complementary approach in flow cytometry and imaging makes it possible to calibrate the biosensor, and to combine the statistical power of cytometry and the resolving power at the tissue level of imaging. A few key words on methods :
Location (adress and meeting point): 1 Avenue de la terrasse, building 21 (ground floor of the Hall), 91198 Gif-sur-Yvette cedex
Dynamic analysis of lipids in cell membranesAIM : Imaging phospholipid dynamics during stimulation of leukocytes Cells : A leukocyte cell line (neutrophil-like) transfected by one or two lipid biosensors Method : 4D video microscopy or TIRF microscopy will be performed in order to follow the localization of the lipid biosensors (phosphoinositide biosensors) at the cell membranes during the stimulation by particle phagocytosis or frustrated phagocytosis. A few key words on methods :
Location (adress and meeting point): 1 Avenue de la terrasse, building 21 (ground floor of the Hall), 91198 Gif-sur-Yvette cedex Measuring bacteria internal pH using PHP biosensor and light microscopyThe legume plants - bacteria symbiosis is characterized by the formation of symbiotic root nodules, in which the bacteria differentiate into nitrogen-fixing bacteroïds. In order to identify novel features of bacteroïd differentiation, we tested a novel genetically encoded ratiometric fluorescent biosensor (pHP, derived from GFP) to estimate intracellular pH. To exploit this biosensor we need to master multiple steps:
A few key words on methods :
Location (adress and meeting point): 1 Avenue de la terrasse, building 21 (ground floor of the Hall), 91198 Gif-sur-Yvette cedex |
| Online user: 2 | Privacy |

|